Software
We have a track record of developing and publicly releasing software for physiological image processing and analysis. Our ethos is to try and make the work we do available for other people to try out and use, seeking to be:
- Transparent: When it comes to algorithms and analysis methods we try hard to make publications as complete as possible. However, it can still be hard for the reader reproduce the method exactly. By providing our code we want to let others have better access to what we have done, compare it to alternative approaches and improve upon it.
- Pragmatic: However good our method may be at solving the problem we designed it for there is a barrier to use because it is not trivial to implement. Since we often try to solve problems that are relevant to people who do not have a highly mathematical or engineering background it seems unfair to make the user re-invent the wheel when we already have the code written.
- Collaborative: Ultimately we want to see research progress and not be held up by making people spend time writing programs that already exist. We also regularly work with people who want to use our methods or even try to improve upon them.
Quantiphyse
Quantiphyse is a GUI viewing and analysis package for biomedical imaging data. As well as generic visualisation and processing functionality, it contains a set of plugins for processing specific kinds of physiological imaging data including ASL, DCE, CEST, DSC and quantitative BOLD.
- Quantiphyse is the main application. Other repositories contain various plugins however documentation for these is kept centrally.
Processing pipelines
These are FSL-based pipelines for processing different types of medical imaging data. They utilise Fabber for the model fitting but also include pre and post processing specific to the data type.
- oxford_asl / basil is the standard FSL based pipeline for processing ASL data.
- oxasl is a new Python-based
pipeline, largely compatible with
oxford_aslbut with some new features and also supporting multiphase and vessel-encoded ASL data. - verbena is a pipeline for processing DSC data.
- hcp-asl is a pipeline for processing ASL data from the Human Connectome Project
Fabber
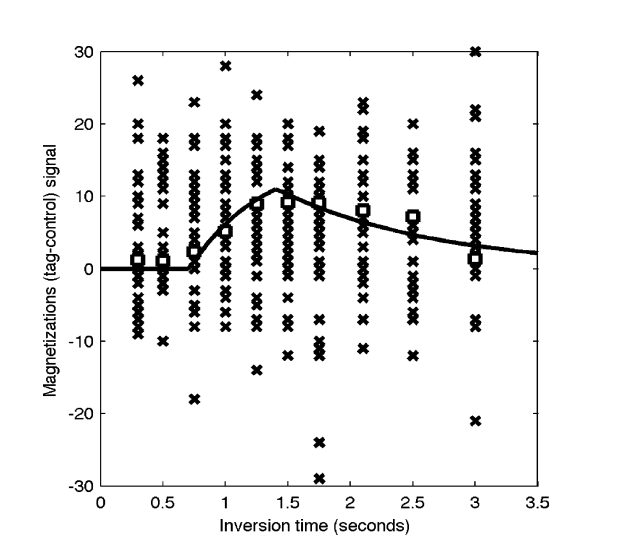
Fabber is a Bayesian model fitting framework which uses the Variational Bayes algorithm to do fast model fitting of nonlinear forward models. It is a fully Bayesian solution allowing the specification of prior information and the production of posterior distributions. The algorithm can be used on any serial data, but we have also designed it to be suitable for serial imaging data.
- Fabber core defines the main model fitting algorithm and contains a few simple generic models for testing and demonstration purposes.
- Fabber ASL models is a set of Fabber models for ASL-MRI data
- Fabber DCE models is a set of Fabber models for DCE-MRI data
- Fabber DSC models is a set of Fabber models for DSC-MRI data
- pyfab is a Python API for Fabber
- fabber_matlab is a simple Matlab interface to Fabber
SVB
SVB is an implementation of Stochastic Variational Bayes for inferring on medical imaging timeseries data. It aims to solve the same problem as Fabber by an alternative means which may provide advantages in some cases.
- SVB is the main implementation currently containing exponential and ASL models
Python libraries
-
Toblerone contains tools for surface based analysis including projection and partial volume estimation. Toblerone can estimate partial volumes across the brain using surface segmentations (for example, those from FreeSurfer and FSL FIRST).
-
Regtricks contains tools for manipulating, combining and applying image transformations. It is not a replacement for image registration tools (eg FSL FLIRT), but it does make working with the output of these tools easier. In particular it enables transformations to be combined within an object oriented interface and applied to data in a single step, avoiding multiple interpolations.
Software Tutorials
-
The FSL course contains a practical session on ASL-MRI processing using the Basil/Oxford ASL toolbox.
-
Neuroimaging Primers is a series of short, accessible textbooks focused on MRI-based neuroimaging. Under
Example Boxesyou can find a set of interactive tutorials on ASL-MRI data processing using the Basil/Oxford ASL toolbox. -
The Variational Bayes Tutorial contains interactive code demonstrating simple implementations of Analytic and Stochastic variational Bayes.
-
Tutorials for DCE, qBOLD and ASL using Quantiphyse are included in the general Quantiphyse documentation